Research
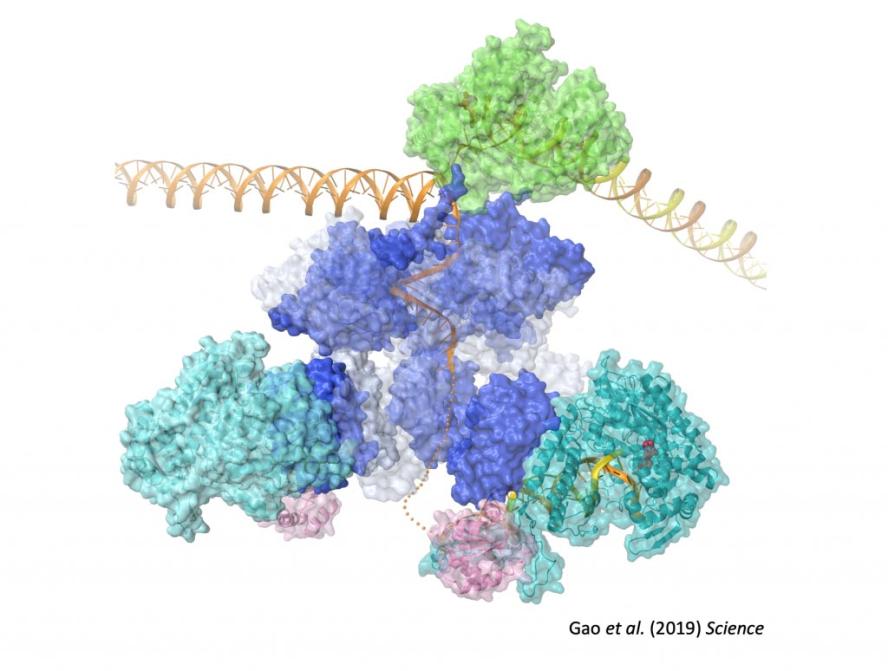
DNA replication from phage to humans
Biochemistry of mitochondrial DNA replication
Mitochondria are essential organelles that produce ATP and are a cellular nexus of metabolism and signaling. Mitochondria are the only organelles in human cells, apart from the nucleus, that have DNA. Mitochondrial DNA (mtDNA) is a double-stranded circle of ~17 kilobase pairs that is replicated by dedicated multiprotein complexes.
The basal replication machinery of human mitochondria is evolutionarily related to the DNA replication machinery of bacteriophage T7. The mitochondrial replicative helicase, TWINKLE, unwinds double-stranded DNA and generates the single-stranded template used by POLγ, the mitochondrial DNA polymerase. The mitochondrial RNA polymerase, POLRMT, synthesizes primers to initiate DNA synthesis, and the mitochondrial single-stranded (ss) DNA-binding protein, mtSSB binds ssDNA regions and stimulates the activity of TWINKLE.
Besides from learning how these fascinating molecular machines work together at a fundamental level, this information will contribute to understanding the causes of human mtDNA diseases, provide the basis to develop novel diagnostic tools and therapeutic avenues for mitochondrial diseases, and help unravel the complex role of mitochondria in human biology.
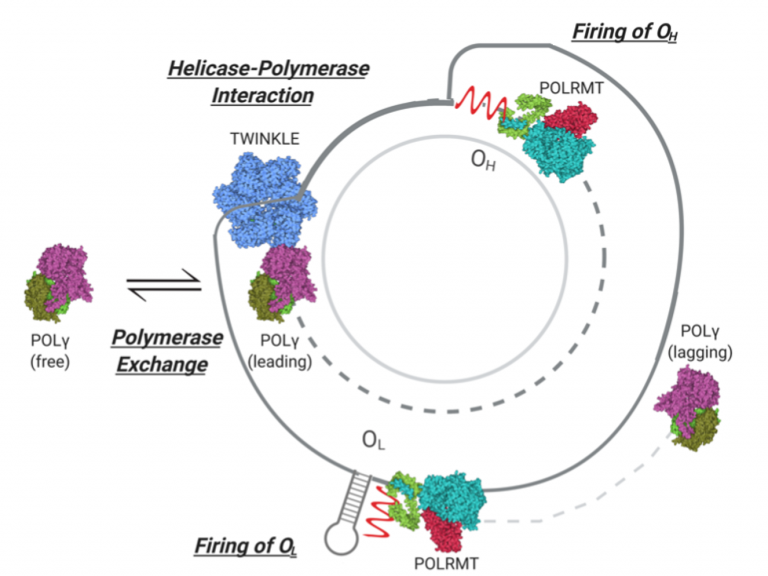
Polymerase exchange during replication
DNA polymerases that are associated with the replisome exchange rapidly with free polymerases in solution. The exchange of polymerases impacts directly the mechanisms of formation of mutations and deletions in mtDNA since both strands of mtDNA are synthesized continuously. Our lab want to understand assembly and exchange dynamics of DNA polymerases during mtDNA elongation, which in turn affects polymerase processivity, fidelity, as well as the response to replisome stalling due to template structure or DNA damage.
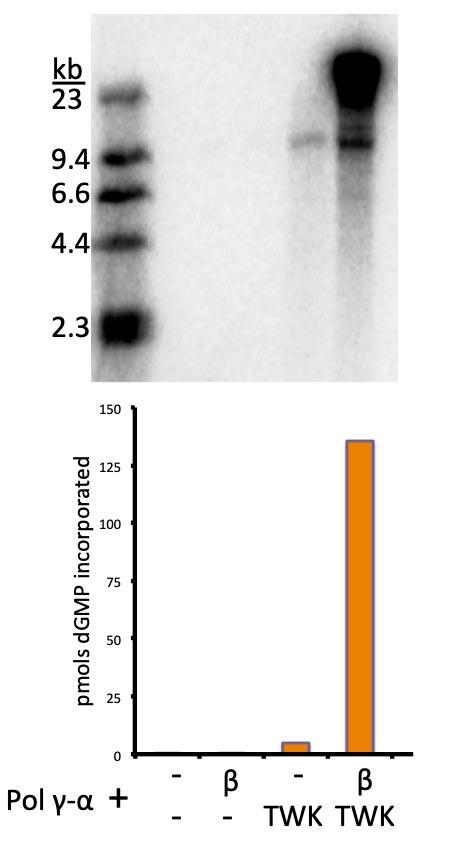
Interaction between TWINKLE and POLγ
Alone, TWINKLE is a weak DNA helicase. Similarly, POLγ cannot utilize dsDNA as a template. But put together, TWINKLE and POLγ form a processive DNA replication machine. Despite multiple lines of indirect evidence suggesting that a specific, functional, interaction between TWINKLE and POLγ occurs, direct proof has been elusive. Our hypothesis is that protein-protein interactions between TWINKLE and POLγ are favored in the context of an advancing replication fork. Our lab will identify the interaction between TWINKLE and POLγ and characterize it biochemically. The interaction between helicase and polymerase which confer processivity may also mediate exchange of replisome components. In addition, other proteins might contribute to regulation of replication elongation. Therefore, understanding the bases for the functional interaction between TWINKLE and POLγ will serve as a platform to understand the effects of the vast list of deleterious mutations in these genes found in patients with mitochondrial disease.
Initiation of mtDNA replication
The initiation of replication of human mtDNA is unusual – each strand has its own origin of replication which fire asynchronously. Replication of the Heavy (H)-strand begins by transcription by POLRMT from a Light (L)-strand promoter. This RNA forms a stable R-loop with template DNA, where transcription is terminated. The molecular details following this event are unclear, but POLγ, in cooperation with TWINKLE, utilizes this RNA as a primer for H-strand synthesis. As the nascent H-strand is elongated, the parental strand is displaced as ssDNA. A stem-loop structure in the displaced strand triggers primer synthesis by POLRMT, and POLγ initiates L-strand synthesis in the opposite direction. A different complement of trans- and cis-acting factors participates in the firing of each replication origin. Despite knowing a number of the players, we still don’t know the rules of the game. The ultimate goal of the lab is the complete in vitro reconstitution of mtDNA replication using purified components, which would allow an unprecedented ability to study this process.
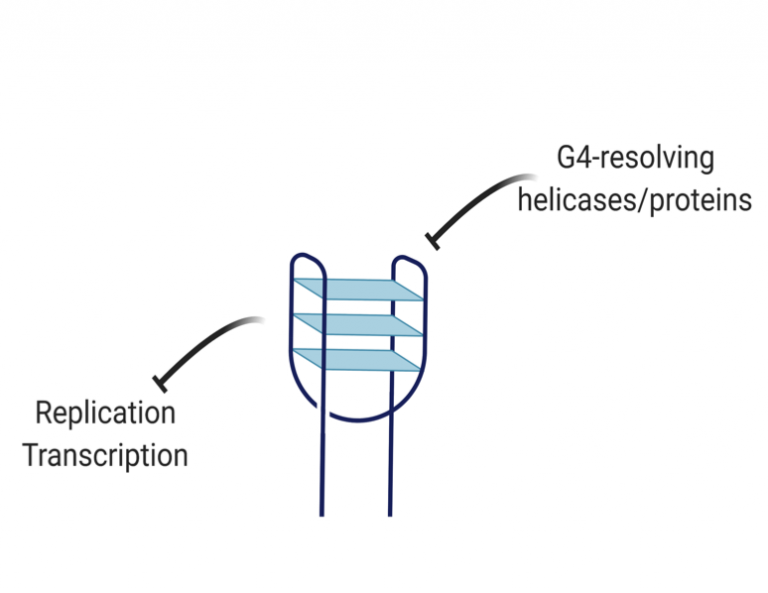
How do mitochondria resolve G-quadruplexes in mtDNA?
G-quadruplexes (G4s) are structures in nucleic acids that form through non-canonical base pairs between guanines and which pose an enormous barrier for DNA replication. Human mtDNA is G-rich and has a 10-fold higher frequency of putative G4-forming regions than nuclear DNA. G4-binding dyes show they are widely present in mtDNA. In addition, G4s map closely to common mutation sites in mtDNA and can block DNA synthesis by POLγ in vitro. Therefore, G4 resolution is likely critical for the maintenance of mtDNA and defects in G4 resolution contribute to mtDNA instability. In line with this notion, there are a number of G4-resolving DNA helicases present in mitochondria whose function is unclear. Our goal is to understand the effects of G4-forming sequences on mtDNA replication and transcription, and how G4-resolving proteins regulate their formation.
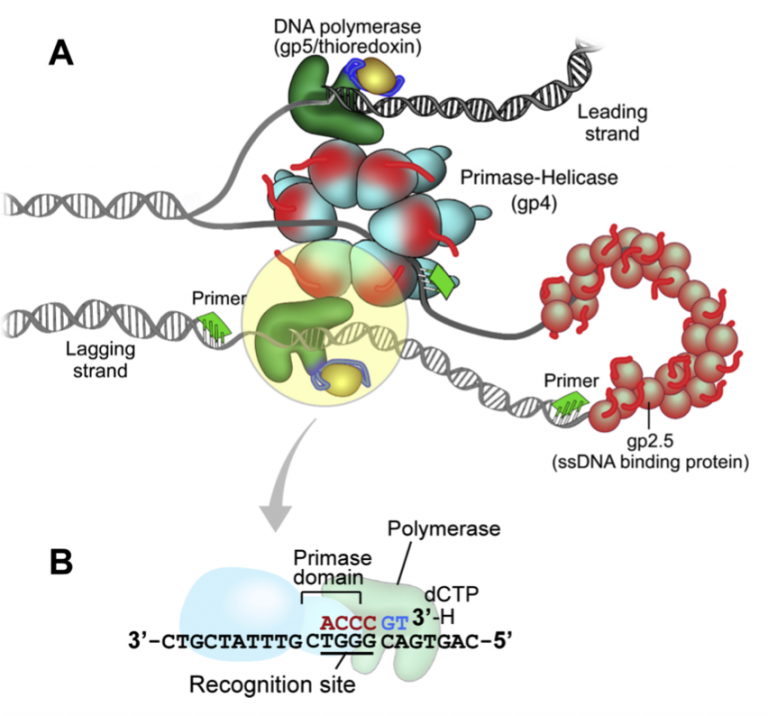
T7 DNA replication
The DNA replication system of bacteriophage T7 is elegantly simple. Only four purified proteins are needed to reconstitute coordinated leading- and lagging-strand DNA synthesis. The T7 chromosome is replicated similarly to more complex bacterial and eukaryotic systems: Initiation of replication occurs at a primary origin, replication is bi-directional, and lagging strand DNA synthesis is dependent on multiple initiation events. Understanding how T7 replication proteins cooperate to replicate DNA will be illustrative to other systems, and this is particularly germane to mtDNA replication.